Research
Understanding molecular mechanisms underpinning biological systems, using tools ranging from atomistic simulations to coarse-grained modelings
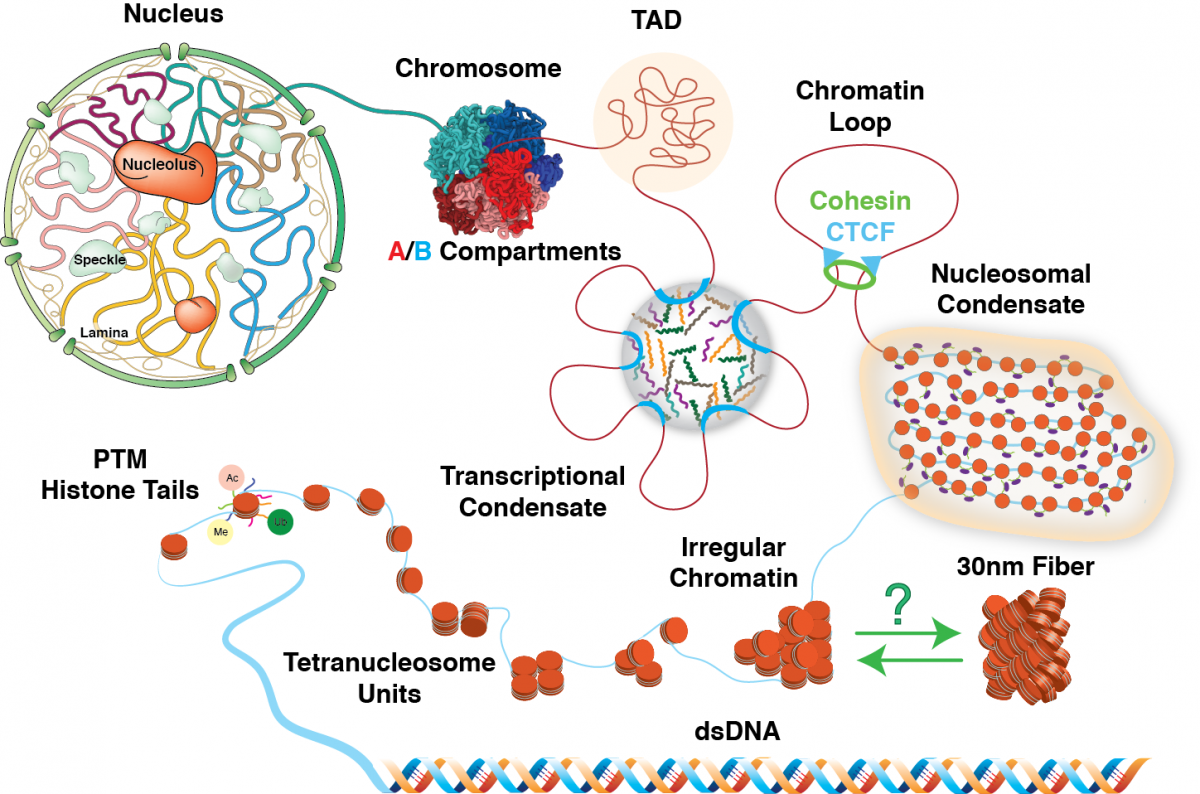 The advance of high-performance computation offers an extraordinary opportunity to study biological macromolecules in silico. My research focuses on models and simulations of biomolecules to understand the underlying physical mechanisms behind various phenomena. Using various computational tools, I aim to capture the interactions among various molecular components quantitatively. Currently, my research focuses on studying the structures and dynamics of chromatin. I am interested in building models to study chromatin fiber, the building blocks of the chromatin system. My research provides a quantitative view of the thermodynamics and kinetics of nucleosomal arrays and how they interact with other cellular components such as epigenetic regulators.
The advance of high-performance computation offers an extraordinary opportunity to study biological macromolecules in silico. My research focuses on models and simulations of biomolecules to understand the underlying physical mechanisms behind various phenomena. Using various computational tools, I aim to capture the interactions among various molecular components quantitatively. Currently, my research focuses on studying the structures and dynamics of chromatin. I am interested in building models to study chromatin fiber, the building blocks of the chromatin system. My research provides a quantitative view of the thermodynamics and kinetics of nucleosomal arrays and how they interact with other cellular components such as epigenetic regulators.
My previous research includes a series topics: 1) exploring the invasion of influenza viruses; 2) building models for the immunological response; 3) developing methods for protein structure prediction; 4) studying proteins implicated in cancer initiation.
During my research experience, I have developed and parameterized models for protein-DNA interactions, accurate and fast predictions of T cell response, and atomistic refinement of protein structures.