Research
Stability and folding pathways of tetranucleosome
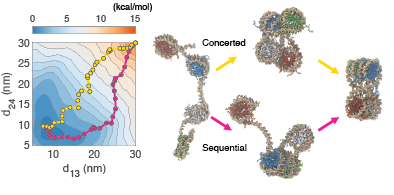
The arrangement of nucleosomes constitutes the building blocks of genome organization. Yet even at the basic level, controversy remains on the folded structure of a series of nucleosomes. Although the long-established picture of ordered 30-nm chromatin fibers was well-characterized in vitro, it disappears in a "sea of nucleosomes" in vivo. Computational modelings offer another avenue to reconcile this uncertainty. We used a near-atomistic chromatin model to study a string of nucleosomes quantitatively. Using enhanced thermodynamic samplings analyzed with deep-learning techniques, we quantitatively characterize the high-dimensional free-energy landscape of a tetranucleosome, the fundamental unit of genomic structure. Our study reveals two pathways connecting the tetranucleosome's native conformation as in the crystal structure to the unfolded states. We find multiple conformations with comparable stabilities to the crystal structure, thus providing evidence for the existence of irregular chromatin structures under a more complicated in vivo environment.
Chromatin fiber under tension and crowding
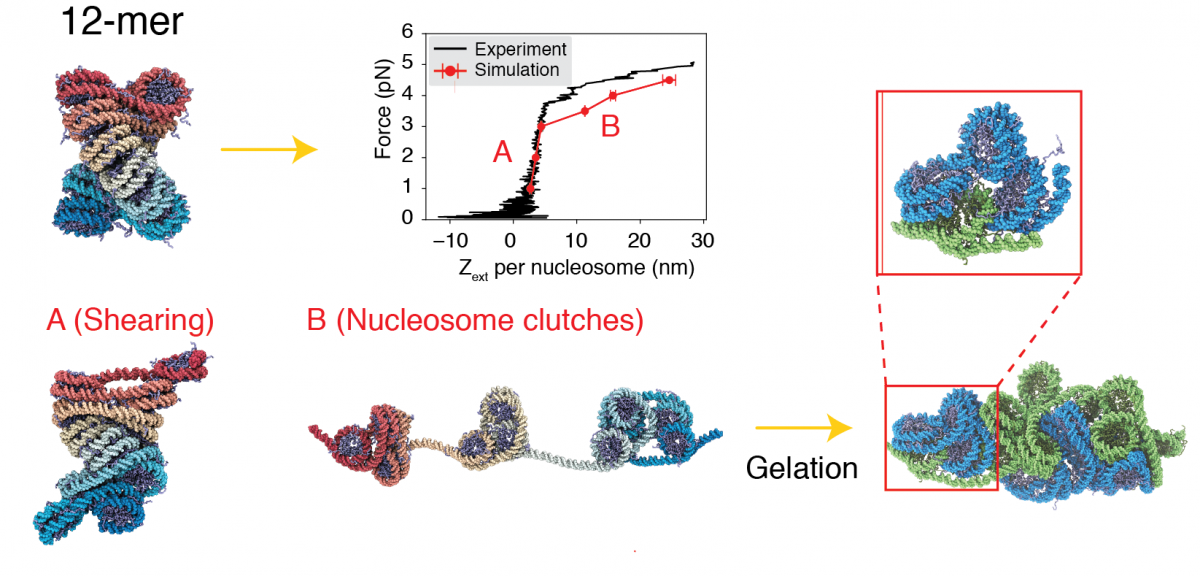 We further utilized this near-atomistic chromatin model to study the impact of tension and crowding on chromatin organization. Our study quantitatively reproduces the response of chromatin under extension force exerted by the single-molecular magnetic tweezer. It further shows that the chromatin breaks into a series of trimeric and tetrameric nucleosome clutches under tension, facilitating the formation of a stable interdigitated configuration by two chromatin chains under a crowded nuclear environment, such as interphase heterochromatin and mitotic chromatin. Our study suggests a viable mechanism for the sol-gel transition of chromatin.
We further utilized this near-atomistic chromatin model to study the impact of tension and crowding on chromatin organization. Our study quantitatively reproduces the response of chromatin under extension force exerted by the single-molecular magnetic tweezer. It further shows that the chromatin breaks into a series of trimeric and tetrameric nucleosome clutches under tension, facilitating the formation of a stable interdigitated configuration by two chromatin chains under a crowded nuclear environment, such as interphase heterochromatin and mitotic chromatin. Our study suggests a viable mechanism for the sol-gel transition of chromatin.
Related publications:
- Shuming Liu†, Xingcheng Lin†, Bin Zhang*, “Chromatin fiber breaks into clutches under tension and crowding”, Nucleic Acids Res., 50, 17, pp 9738-9747, (2022)
- Xingcheng Lin†, Yifeng Qi†, Andrew Latham, Bin Zhang*, "Multiscale Modeling of Genome Organization with Maximum Entropy Optimization" J. Chem. Phys., 155, 010901, (2021)
- Xinqiang Ding†, Xingcheng Lin†, Bin Zhang*, “Stability and folding pathways of tetra-nucleosome from six-dimensional free energy surface”, Nature Communications, 12:1091, (2021)