Research
Epigenetic regulation by Polycomb group proteins
Multicellular organisms develop various kinds of cell identities that are maintained by epigenetic signals. How epigenetic signals were inherited and maintained through cell division and differentiation is largely unclear. One typical means of establishing epigenetic signals is through posttranslational modification of nucleosomal histones. The histone modifications were established and transmitted through depositing modification marks on new histones by a series of protein complexes called epigenetic regulators. The Polycomb group (PcG) proteins are a group of proteins constituting essential members of chromatin regulators. The PcG complexes include two major families: Polycomb repressive complexes 1 and 2 (PRC1 and PRC2). The PcG complexes are responsible for silencing gene expressions through catalytic deposition of histone marks or independently reorganization of chromatin structure. PcG complexes also closely interact with other cellular components such as other DNA-binding proteins and RNAs to regulate gene expression.
1) Methyltransferase activity of PRC2 through binding nonadjacent nucleosomes
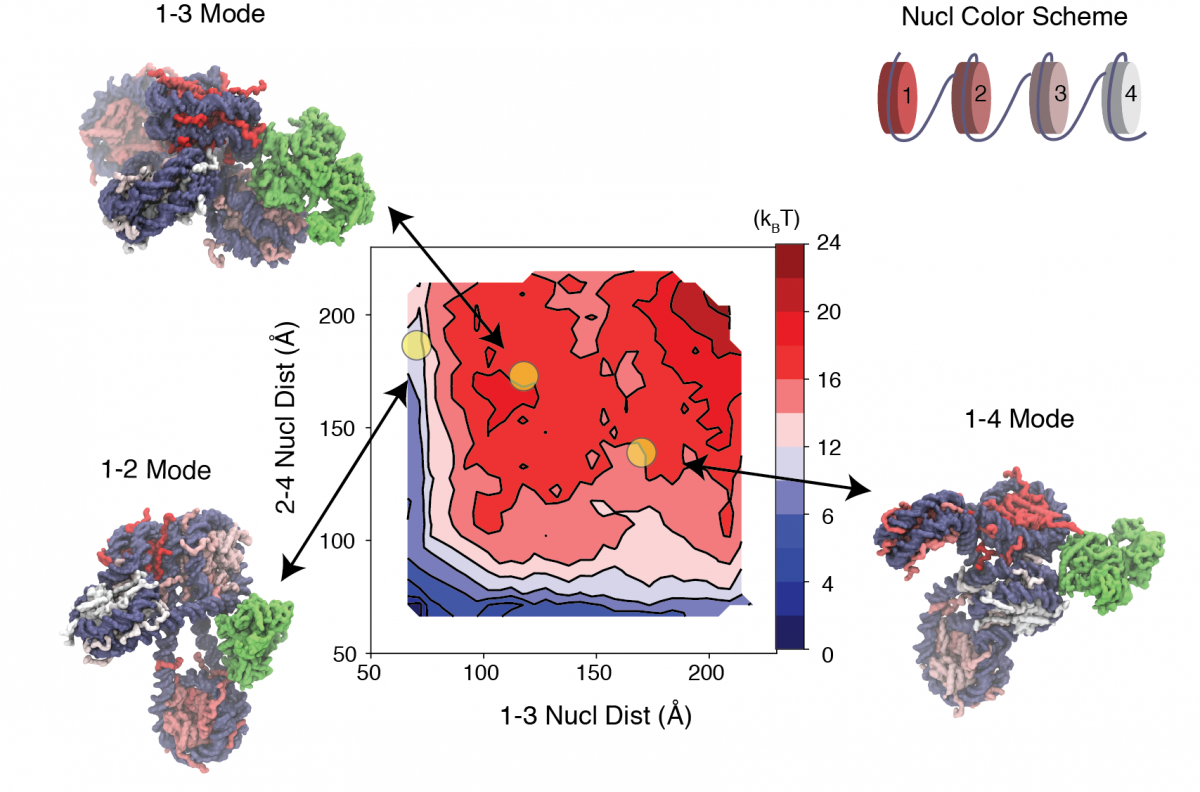
In Collaboration with Prof. Shixin Liu's lab, we quantitatively characterize the interactions between PRC2 and polynucleosome arrays. Through a combination of single-molecule force spectroscopy and computational modeling, we find PRC2 stably binds non-adjacent nucleosomes. Our work suggests an efficient way for PRC2 to spread histone marks in a non-linear fashion, i.e., PRC2 engages non-neighboring nucleosomes to maintain condensed chromatin.
2) Cooperative looping of DNA by PRC2
PRC2 is a large protein complex consisting of multiple components. By a combination of its components, different oligomeric states PRC2 assumes different molecular functions. Due to its size and heterogeneity, the complete structure of PRC2 is unclear, though each individual component of PRC2 has been partially solved by cryogenic electron microscopy and X-ray crystallography. We computationally modeled the complete oligomeric core structures of PRC2 in both monomeric and dimeric state, based on existing experimental data, and studied how PRC2 binds and loops a double-strand DNA (dsDNA) in molecular details. The PRC2-DNA interaction was believed to be an essential factor for PRC2 to compact chromatin structure.
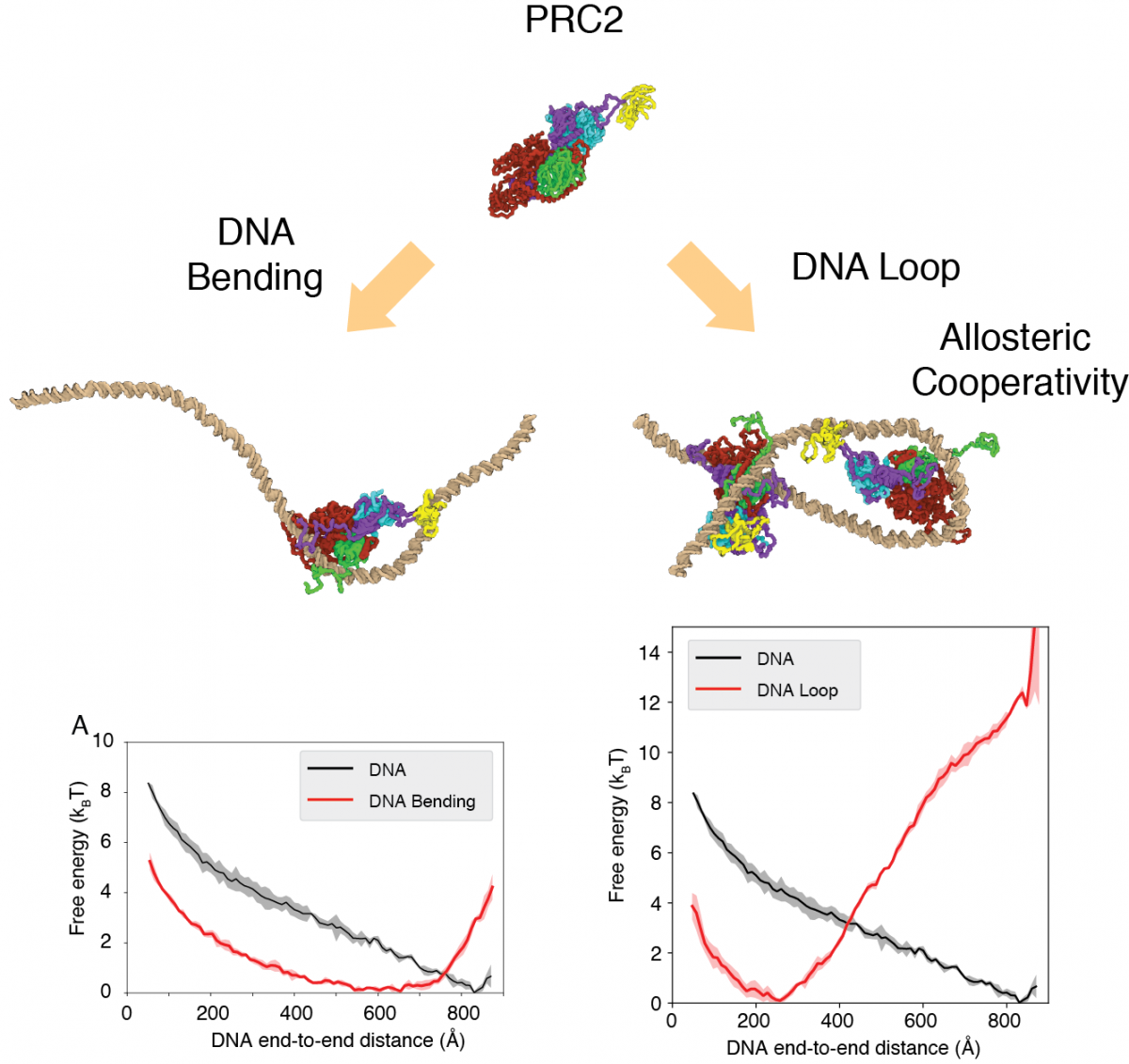 We parameterized a coarse-grained protein-DNA force field which better captures the binding affinity between DNA and known DNA-binding proteins. We study the PRC2-DNA interactions through molecular dynamics simulations combined with enhanced sampling techniques. We find multiple PRC2s cooperatively loops a dsDNA. Such cooperativity does not involve direct protein-protein interactions and is mediated by the DNA. This loop can be further stabilized by the exposed C2 domain of the core PRC2 structure. The cooperative looping serves as an efficient means for multiple PRC2s to condense chromatin structure, independent of its methyltransferase activity. The newly parameterized protein-DNA force field can be applied in simulations to understand the interactions of other protein-DNA complexes.
We parameterized a coarse-grained protein-DNA force field which better captures the binding affinity between DNA and known DNA-binding proteins. We study the PRC2-DNA interactions through molecular dynamics simulations combined with enhanced sampling techniques. We find multiple PRC2s cooperatively loops a dsDNA. Such cooperativity does not involve direct protein-protein interactions and is mediated by the DNA. This loop can be further stabilized by the exposed C2 domain of the core PRC2 structure. The cooperative looping serves as an efficient means for multiple PRC2s to condense chromatin structure, independent of its methyltransferase activity. The newly parameterized protein-DNA force field can be applied in simulations to understand the interactions of other protein-DNA complexes.
Related Publications:
- Xingcheng Lin, Rachel Leicher, Shixin Liu, Bin Zhang*, "Cooperative DNA Looping by PRC2 Complexes via Allosteric Communication", Nucleic Acids Res., 49, 11, pp 6238-6248, (2021)
- Rachel Leicher, Eva J. Ge†, Xingcheng Lin†, Matthew J. Reynolds†, Thomas Walz, Bin Zhang*, Tom W. Muir, Shixin Liu*, "Single-molecule and in silico dissection of the interaction between Polycomb repressive complex 2 and chromatin", Proc. Natl. Acad. Sci. USA, 117, pp 30465-30475, (2020)